Automatic classification¶
EXERCISE: Basic classification¶
The file you need is in the basic-classification folder. Follow the instructions below.
This example introduces ‘defined classes’ and automatic classification. The example involves classification of different ubiquitin ligase complexes. It is based on a subset of the gene ontology (GO) http://purl.obolibrary.org/obo/go.owl with some classes removed for teaching purposes.
Constructs:
- and (intersection)
- equivalence (logical definitions)
- existential restrictions (e.g. part_of some)
Background knowledge for non-GO people:
GO includes pre-composed grouping classes such as ‘chromosomal part’ and ‘nuclear part’.
PART 1: Adding classes and automatically classifying them
- Open basic-classification/ubiq-ligase-complex.owl
- Navigate to ‘ubiquitin ligase complex’
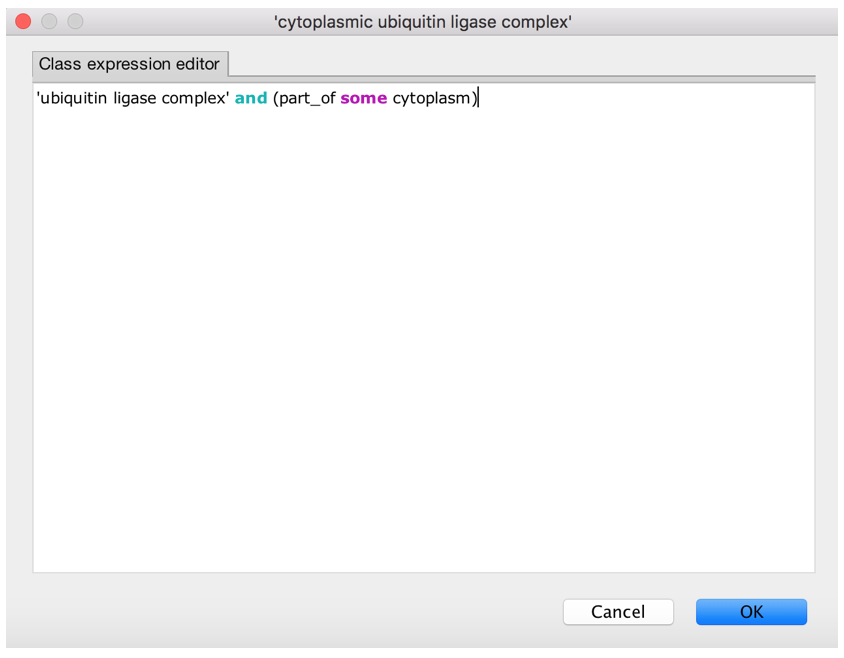
- Add a subclass of ‘ubiquitin ligase complex’ called ‘cytoplasmic ubiquitin ligase complex’
- NOTE: do this *directly* under the ‘ubiquitin ligase complex’ class, don’t move things around!
- NOTE: this class already exists in the main GO, but it has been removed for this tutorial example
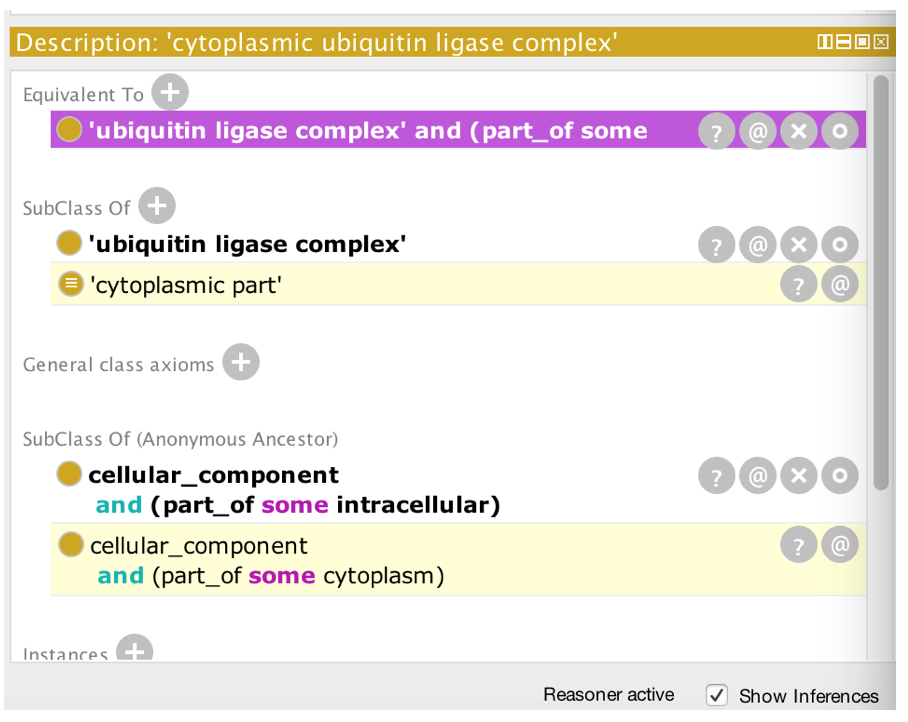
- Give it a logical definition (equivalence axiom)
- Synchronize the reasoner
- Find the class you made under ‘Class hierarchy (inferred)’
- You should see ‘cytoplasmic part’ is now inferred to be a parent class of ‘cytoplasmic ubiquitin ligase complex’.
PART 2: Another example
- Do the same for ‘nuclear ubiquitin ligase complex’ – create the class and add the equivalence axiom.
- Synchronize the reasoner.
- You should see ‘nuclear ubiquitin ligase complex’ is inferred to be a child of ‘nuclear part’.
BONUS:
- Remove the classes you have created.
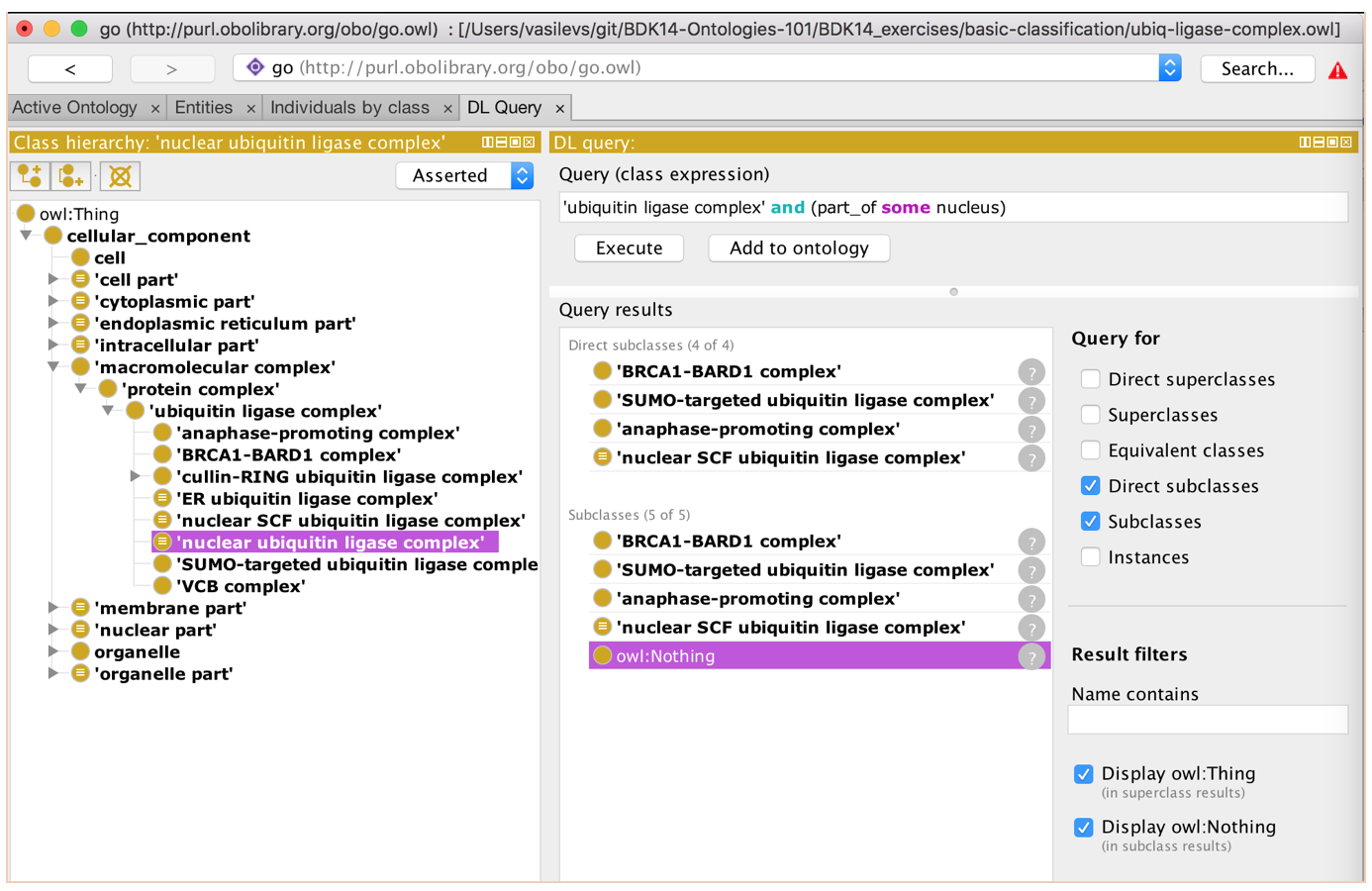
- Find all ‘ubiquitin ligase complex’ classes whose instances are in a nucleus in the DL query tab
- Make the class directly from here